Note
Click here to download the full example code
Compute markers used for DOC-Forest recipe¶
Here we compute the markers from previously computed markers as published [1].
For simplicity, we only compute scalars using a trimmed mean (80%) accross epochs and the mean across channels.
References¶
| [1] | Engemann D.A.*, Raimondo F.*, King JR., Rohaut B., Louppe G., Faugeras F., Annen J., Cassol H., Gosseries O., Fernandez-Slezak D., Laureys S., Naccache L., Dehaene S. and Sitt J.D. (2018). Robust EEG-based cross-site and cross-protocol classification of states of consciousness. Brain. doi:10.1093/brain/awy251 |
# Authors: Denis A. Engemann <denis.engemann@gmail.com>
# Federico Raimondo <federaimondo@gmail.com>
import numpy as np
from scipy.stats import trim_mean
import os.path as op
import mne
from nice import read_markers
import matplotlib.pyplot as plt
from matplotlib.path import Path
from matplotlib.patches import PathPatch
from mpl_toolkits.axes_grid1 import make_axes_locatable
import seaborn as sns
sns.set_color_codes()
def trim_mean80(a, axis=0): # noqa
return trim_mean(a, proportiontocut=.1, axis=axis)
def entropy(a, axis=0): # noqa
return -np.nansum(a * np.log(a), axis=axis) / np.log(a.shape[axis])
fname = 'data/JSXXX-markers.hdf5'
if not op.exists(fname):
raise ValueError('Please run compute_doc_forest_markers.py example first')
fc = read_markers(fname)
Out:
616 matching events found
No baseline correction applied
Not setting metadata
Created an SSP operator (subspace dimension = 2)
2 projection items activated
0 bad epochs dropped
Set regions of interest
For some markers we do not want to use all channels. We therefore supply selections of channels for some markers.
scalp_roi = np.arange(224)
non_scalp = np.arange(224, 256)
cnv_roi = np.array([5, 6, 13, 14, 15, 21, 22])
mmn_roi = np.array([5, 6, 8, 13, 14, 15, 21, 22, 44, 80, 131, 185])
p3b_roi = np.array([8, 44, 80, 99, 100, 109, 118, 127, 128, 131, 185])
p3a_roi = np.array([5, 6, 8, 13, 14, 15, 21, 22, 44, 80, 131, 185])
Set reduction functions
We want delineate different features from each marker. We therefore summarize each marker over epochs and channels. Here we only compute the mean over epochs and channels.
channels_fun = np.mean # function to summarize channels
epochs_fun = trim_mean80 # robust mean to summarize epochs
# For each class of marker we can specify how the reductions have to be
# computed. Each class therefore gets an entry in the `reduction_params`.
# This has to be a dictionary with the keys `reduction_func` and `picks`.
# The first key is a list and can be read as follows: for each reduction,
# sequentially apply `function` over `axis` and then pass the output to the
# next step. For the first example below, we first compute the mean over
# epochs, then the mean over channelsm, and finally, the sum over frequencies.
# While doing so, only consider the channels in `picks`.
# We could also specificy which epochs to use by setting `epochs`.
# We will do this for each class of markers.
reduction_params = {}
reduction_params['PowerSpectralDensity'] = {
'reduction_func':
[{'axis': 'epochs', 'function': epochs_fun},
{'axis': 'channels', 'function': channels_fun},
{'axis': 'frequency', 'function': np.sum}],
'picks': {
'epochs': None,
'channels': scalp_roi}}
reduction_params['PowerSpectralDensity/summary_se'] = {
'reduction_func':
[{'axis': 'frequency', 'function': entropy},
{'axis': 'epochs', 'function': np.mean},
{'axis': 'channels', 'function': channels_fun}],
'picks': {
'epochs': None,
'channels': scalp_roi}}
reduction_params['PowerSpectralDensitySummary'] = {
'reduction_func':
[{'axis': 'epochs', 'function': epochs_fun},
{'axis': 'channels', 'function': channels_fun}],
'picks': {
'epochs': None,
'channels': scalp_roi}}
reduction_params['PermutationEntropy'] = {
'reduction_func':
[{'axis': 'epochs', 'function': epochs_fun},
{'axis': 'channels', 'function': channels_fun}],
'picks': {
'epochs': None,
'channels': scalp_roi}}
reduction_params['SymbolicMutualInformation'] = {
'reduction_func':
[{'axis': 'epochs', 'function': epochs_fun},
{'axis': 'channels_y', 'function': np.median},
{'axis': 'channels', 'function': channels_fun}],
'picks': {
'epochs': None,
'channels_y': scalp_roi,
'channels': scalp_roi}}
reduction_params['KolmogorovComplexity'] = {
'reduction_func':
[{'axis': 'epochs', 'function': epochs_fun},
{'axis': 'channels', 'function': channels_fun}],
'picks': {
'epochs': None,
'channels': scalp_roi}}
reduction_params['ContingentNegativeVariation'] = {
'reduction_func':
[{'axis': 'epochs', 'function': epochs_fun},
{'axis': 'channels', 'function': channels_fun}],
'picks': {
'epochs': None,
'channels': cnv_roi}}
reduction_params['TimeLockedTopography'] = {
'reduction_func':
[{'axis': 'epochs', 'function': epochs_fun},
{'axis': 'channels', 'function': channels_fun},
{'axis': 'times', 'function': np.mean}],
'picks': {
'epochs': None,
'channels': scalp_roi,
'times': None}}
reduction_params['TimeLockedContrast'] = {
'reduction_func':
[{'axis': 'epochs', 'function': epochs_fun},
{'axis': 'channels', 'function': channels_fun},
{'axis': 'times', 'function': np.mean}],
'picks': {
'epochs': None,
'channels': scalp_roi,
'times': None}}
reduction_params['TimeLockedContrast/mmn'] = {
'reduction_func':
[{'axis': 'epochs', 'function': epochs_fun},
{'axis': 'channels', 'function': channels_fun},
{'axis': 'times', 'function': np.mean}],
'picks': {
'epochs': None,
'channels': mmn_roi,
'times': None}}
reduction_params['TimeLockedContrast/p3b'] = {
'reduction_func':
[{'axis': 'epochs', 'function': epochs_fun},
{'axis': 'channels', 'function': channels_fun},
{'axis': 'times', 'function': np.mean}],
'picks': {
'epochs': None,
'channels': p3b_roi,
'times': None}}
reduction_params['TimeLockedContrast/p3a'] = {
'reduction_func':
[{'axis': 'epochs', 'function': epochs_fun},
{'axis': 'channels', 'function': channels_fun},
{'axis': 'times', 'function': np.mean}],
'picks': {
'epochs': None,
'channels': p3a_roi,
'times': None}}
Actually compute reductions
Now we can summarize the markers either into scalars (1 marker, 1 value) or topos (1 marker, n_channels values).
scalars = fc.reduce_to_scalar(reduction_params)
topos = fc.reduce_to_topo(reduction_params)
# Those are numpy arrays.
print('%i markers' % scalars.shape)
print('%i markers, %i channels' % topos.shape)
Out:
Reducing to scalars
Reducing nice/marker/PowerSpectralDensity/delta
Reduction order for nice/marker/PowerSpectralDensity/delta: ['epochs', 'channels', 'frequency']
Reducing nice/marker/PowerSpectralDensity/deltan
Reduction order for nice/marker/PowerSpectralDensity/deltan: ['epochs', 'channels', 'frequency']
Reducing nice/marker/PowerSpectralDensity/theta
Reduction order for nice/marker/PowerSpectralDensity/theta: ['epochs', 'channels', 'frequency']
Reducing nice/marker/PowerSpectralDensity/thetan
Reduction order for nice/marker/PowerSpectralDensity/thetan: ['epochs', 'channels', 'frequency']
Reducing nice/marker/PowerSpectralDensity/alpha
Reduction order for nice/marker/PowerSpectralDensity/alpha: ['epochs', 'channels', 'frequency']
Reducing nice/marker/PowerSpectralDensity/alphan
Reduction order for nice/marker/PowerSpectralDensity/alphan: ['epochs', 'channels', 'frequency']
Reducing nice/marker/PowerSpectralDensity/beta
Reduction order for nice/marker/PowerSpectralDensity/beta: ['epochs', 'channels', 'frequency']
Reducing nice/marker/PowerSpectralDensity/betan
Reduction order for nice/marker/PowerSpectralDensity/betan: ['epochs', 'channels', 'frequency']
Reducing nice/marker/PowerSpectralDensity/gamma
Reduction order for nice/marker/PowerSpectralDensity/gamma: ['epochs', 'channels', 'frequency']
Reducing nice/marker/PowerSpectralDensity/gamman
Reduction order for nice/marker/PowerSpectralDensity/gamman: ['epochs', 'channels', 'frequency']
Reducing nice/marker/PowerSpectralDensity/summary_se
Reduction order for nice/marker/PowerSpectralDensity/summary_se: ['frequency', 'epochs', 'channels']
Reducing nice/marker/PowerSpectralDensitySummary/summary_msf
Reduction order for nice/marker/PowerSpectralDensitySummary/summary_msf: ['epochs', 'channels']
Reducing nice/marker/PowerSpectralDensitySummary/summary_sef90
Reduction order for nice/marker/PowerSpectralDensitySummary/summary_sef90: ['epochs', 'channels']
Reducing nice/marker/PowerSpectralDensitySummary/summary_sef95
Reduction order for nice/marker/PowerSpectralDensitySummary/summary_sef95: ['epochs', 'channels']
Reducing nice/marker/PermutationEntropy/default
Reduction order for nice/marker/PermutationEntropy/default: ['epochs', 'channels']
Reducing nice/marker/SymbolicMutualInformation/weighted
Reduction order for nice/marker/SymbolicMutualInformation/weighted: ['epochs', 'channels_y', 'channels']
Reducing nice/marker/KolmogorovComplexity/default
Reduction order for nice/marker/KolmogorovComplexity/default: ['epochs', 'channels']
Reducing nice/marker/ContingentNegativeVariation/default
Reduction order for nice/marker/ContingentNegativeVariation/default: ['epochs', 'channels']
Reducing nice/marker/TimeLockedTopography/p1
Reduction order for nice/marker/TimeLockedTopography/p1: ['epochs', 'channels', 'times']
Reducing nice/marker/TimeLockedTopography/p3a
Reduction order for nice/marker/TimeLockedTopography/p3a: ['epochs', 'channels', 'times']
Reducing nice/marker/TimeLockedTopography/p3b
Reduction order for nice/marker/TimeLockedTopography/p3b: ['epochs', 'channels', 'times']
Reducing nice/marker/TimeLockedContrast/LSGS-LDGD
Reduction order for nice/marker/TimeLockedTopography/default: ['epochs', 'channels', 'times']
Reduction order for nice/marker/TimeLockedTopography/default: ['epochs', 'channels', 'times']
Reducing nice/marker/TimeLockedContrast/LSGD-LDGS
Reduction order for nice/marker/TimeLockedTopography/default: ['epochs', 'channels', 'times']
Reduction order for nice/marker/TimeLockedTopography/default: ['epochs', 'channels', 'times']
Reducing nice/marker/TimeLockedContrast/LD-LS
Reduction order for nice/marker/TimeLockedTopography/default: ['epochs', 'channels', 'times']
Reduction order for nice/marker/TimeLockedTopography/default: ['epochs', 'channels', 'times']
Reducing nice/marker/TimeLockedContrast/mmn
Reduction order for nice/marker/TimeLockedTopography/default: ['epochs', 'channels', 'times']
Reduction order for nice/marker/TimeLockedTopography/default: ['epochs', 'channels', 'times']
Reducing nice/marker/TimeLockedContrast/p3a
Reduction order for nice/marker/TimeLockedTopography/default: ['epochs', 'channels', 'times']
Reduction order for nice/marker/TimeLockedTopography/default: ['epochs', 'channels', 'times']
Reducing nice/marker/TimeLockedContrast/GD-GS
Reduction order for nice/marker/TimeLockedTopography/default: ['epochs', 'channels', 'times']
Reduction order for nice/marker/TimeLockedTopography/default: ['epochs', 'channels', 'times']
Reducing nice/marker/TimeLockedContrast/p3b
Reduction order for nice/marker/TimeLockedTopography/default: ['epochs', 'channels', 'times']
Reduction order for nice/marker/TimeLockedTopography/default: ['epochs', 'channels', 'times']
Reducing to topographies
Reducing nice/marker/PowerSpectralDensity/delta
Reduction order for nice/marker/PowerSpectralDensity/delta: ['epochs', 'frequency']
Reducing nice/marker/PowerSpectralDensity/deltan
Reduction order for nice/marker/PowerSpectralDensity/deltan: ['epochs', 'frequency']
Reducing nice/marker/PowerSpectralDensity/theta
Reduction order for nice/marker/PowerSpectralDensity/theta: ['epochs', 'frequency']
Reducing nice/marker/PowerSpectralDensity/thetan
Reduction order for nice/marker/PowerSpectralDensity/thetan: ['epochs', 'frequency']
Reducing nice/marker/PowerSpectralDensity/alpha
Reduction order for nice/marker/PowerSpectralDensity/alpha: ['epochs', 'frequency']
Reducing nice/marker/PowerSpectralDensity/alphan
Reduction order for nice/marker/PowerSpectralDensity/alphan: ['epochs', 'frequency']
Reducing nice/marker/PowerSpectralDensity/beta
Reduction order for nice/marker/PowerSpectralDensity/beta: ['epochs', 'frequency']
Reducing nice/marker/PowerSpectralDensity/betan
Reduction order for nice/marker/PowerSpectralDensity/betan: ['epochs', 'frequency']
Reducing nice/marker/PowerSpectralDensity/gamma
Reduction order for nice/marker/PowerSpectralDensity/gamma: ['epochs', 'frequency']
Reducing nice/marker/PowerSpectralDensity/gamman
Reduction order for nice/marker/PowerSpectralDensity/gamman: ['epochs', 'frequency']
Reducing nice/marker/PowerSpectralDensity/summary_se
Reduction order for nice/marker/PowerSpectralDensity/summary_se: ['frequency', 'epochs']
Reducing nice/marker/PowerSpectralDensitySummary/summary_msf
Reduction order for nice/marker/PowerSpectralDensitySummary/summary_msf: ['epochs']
Reducing nice/marker/PowerSpectralDensitySummary/summary_sef90
Reduction order for nice/marker/PowerSpectralDensitySummary/summary_sef90: ['epochs']
Reducing nice/marker/PowerSpectralDensitySummary/summary_sef95
Reduction order for nice/marker/PowerSpectralDensitySummary/summary_sef95: ['epochs']
Reducing nice/marker/PermutationEntropy/default
Reduction order for nice/marker/PermutationEntropy/default: ['epochs']
Reducing nice/marker/SymbolicMutualInformation/weighted
Reduction order for nice/marker/SymbolicMutualInformation/weighted: ['epochs', 'channels_y']
Reducing nice/marker/KolmogorovComplexity/default
Reduction order for nice/marker/KolmogorovComplexity/default: ['epochs']
Reducing nice/marker/ContingentNegativeVariation/default
Reduction order for nice/marker/ContingentNegativeVariation/default: ['epochs']
Reducing nice/marker/TimeLockedTopography/p1
Reduction order for nice/marker/TimeLockedTopography/p1: ['epochs', 'times']
Reducing nice/marker/TimeLockedTopography/p3a
Reduction order for nice/marker/TimeLockedTopography/p3a: ['epochs', 'times']
Reducing nice/marker/TimeLockedTopography/p3b
Reduction order for nice/marker/TimeLockedTopography/p3b: ['epochs', 'times']
Reducing nice/marker/TimeLockedContrast/LSGS-LDGD
Reduction order for nice/marker/TimeLockedTopography/default: ['epochs', 'times']
Reduction order for nice/marker/TimeLockedTopography/default: ['epochs', 'times']
Reducing nice/marker/TimeLockedContrast/LSGD-LDGS
Reduction order for nice/marker/TimeLockedTopography/default: ['epochs', 'times']
Reduction order for nice/marker/TimeLockedTopography/default: ['epochs', 'times']
Reducing nice/marker/TimeLockedContrast/LD-LS
Reduction order for nice/marker/TimeLockedTopography/default: ['epochs', 'times']
Reduction order for nice/marker/TimeLockedTopography/default: ['epochs', 'times']
Reducing nice/marker/TimeLockedContrast/mmn
Reduction order for nice/marker/TimeLockedTopography/default: ['epochs', 'times']
Reduction order for nice/marker/TimeLockedTopography/default: ['epochs', 'times']
Reducing nice/marker/TimeLockedContrast/p3a
Reduction order for nice/marker/TimeLockedTopography/default: ['epochs', 'times']
Reduction order for nice/marker/TimeLockedTopography/default: ['epochs', 'times']
Reducing nice/marker/TimeLockedContrast/GD-GS
Reduction order for nice/marker/TimeLockedTopography/default: ['epochs', 'times']
Reduction order for nice/marker/TimeLockedTopography/default: ['epochs', 'times']
Reducing nice/marker/TimeLockedContrast/p3b
Reduction order for nice/marker/TimeLockedTopography/default: ['epochs', 'times']
Reduction order for nice/marker/TimeLockedTopography/default: ['epochs', 'times']
28 markers
28 markers, 256 channels
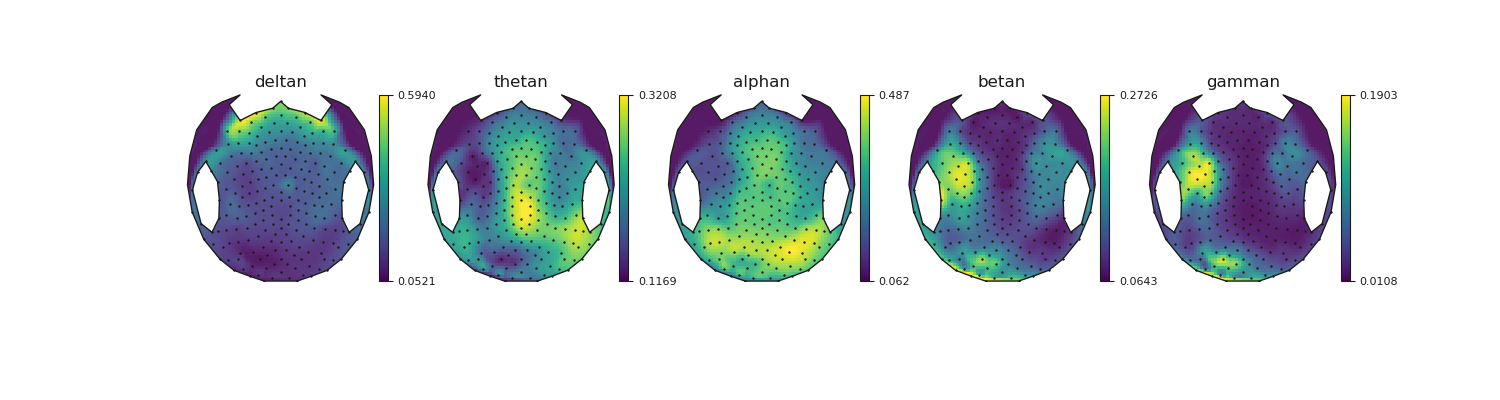
Plot a few markers
# Let's create convenient names from the marker keys.
to_plot = ['nice/marker/PowerSpectralDensity/deltan',
'nice/marker/PowerSpectralDensity/thetan',
'nice/marker/PowerSpectralDensity/alphan',
'nice/marker/PowerSpectralDensity/betan',
'nice/marker/PowerSpectralDensity/gamman']
idx = [list(fc.keys()).index(x) for x in to_plot]
names = [x.split('/')[-1] for x in to_plot]
topos_to_plot = topos[idx]
# Prepare fancy EGI plot with nicer outline.
montage = mne.channels.read_montage('GSN-HydroCel-256')
ch_names = ['E{}'.format(i) for i in range(1, 257)]
info = mne.create_info(ch_names, 1, ch_types='eeg', montage=montage)
layout = mne.channels.make_eeg_layout(info)
pos = layout.pos[:, :2]
_egi256_outlines = {
'ear1': np.array([190, 191, 201, 209, 218, 217, 216, 208, 200, 190]),
'ear2': np.array([81, 72, 66, 67, 68, 73, 82, 92, 91, 81]),
'outer': np.array([9, 17, 24, 30, 31, 36, 45, 243, 240, 241, 242, 246, 250,
255, 90, 101, 110, 119, 132, 144, 164, 173, 186, 198,
207, 215, 228, 232, 236, 239, 238, 237, 233, 9]),
}
outlines = {}
codes = []
vertices = []
for k, v in _egi256_outlines.items():
t_verts = pos[v, :]
outlines[k] = (t_verts[:, 0], t_verts[:, 1])
t_codes = 2 * np.ones(v.shape[0])
t_codes[0] = 1
codes.append(t_codes)
vertices.append(t_verts)
vertices = np.concatenate(vertices, axis=0)
codes = np.concatenate(codes, axis=0)
path = Path(vertices=vertices, codes=codes)
def patch(): # noqa
return PathPatch(path, color='white', alpha=0.1)
outlines['mask_pos'] = outlines['outer']
outlines['patch'] = patch
pos = layout.pos[:, :2]
mask = np.in1d(np.arange(len(pos)), scalp_roi)
mask_params = dict(marker='+', markerfacecolor='k', markeredgecolor='k',
linewidth=0, markersize=1)
cmap = 'viridis'
n_axes = len(names)
fig_kwargs = dict(figsize=(3 * n_axes, 4))
fig, axes = plt.subplots(1, n_axes, **fig_kwargs)
for ax, name, topo in zip(axes, names, topos_to_plot):
vmin = np.nanmin(topo[scalp_roi])
vmax = np.nanmax(topo[scalp_roi])
topo[non_scalp] = vmin
nan_idx = np.isnan(topo)
im, _ = mne.viz.topomap.plot_topomap(
topo[~nan_idx], pos[~nan_idx], vmin=vmin, vmax=vmax, axes=ax,
cmap=cmap, image_interp='nearest', outlines=outlines, sensors=False,
mask=mask, mask_params=mask_params, contours=0)
ax.set_title(name)
divider = make_axes_locatable(ax)
cax = divider.append_axes("right", size="5%", pad=0.05)
cbar = plt.colorbar(im, cax=cax, ticks=(vmin, vmax))
cbar.ax.tick_params(labelsize=8)
plt.show()
Total running time of the script: ( 3 minutes 3.416 seconds)