Note
Click here to download the full example code
Compute markers used for publication¶
Here we compute the markers used for the diagnosis of DOC patients [1] for an EGI recording from a control subject.
References¶
| [1] | Engemann D.A.*, Raimondo F.*, King JR., Rohaut B., Louppe G., Faugeras F., Annen J., Cassol H., Gosseries O., Fernandez-Slezak D., Laureys S., Naccache L., Dehaene S. and Sitt J.D. (2018). Robust EEG-based cross-site and cross-protocol classification of states of consciousness. Brain. doi:10.1093/brain/awy251 |
# Authors: Denis A. Engemann <denis.engemann@gmail.com>
# Federico Raimondo <federaimondo@gmail.com>
import os.path as op
import mne
import numpy as np
import matplotlib.pyplot as plt
from nice import Markers
from nice.markers import (PowerSpectralDensity,
KolmogorovComplexity,
PermutationEntropy,
SymbolicMutualInformation,
PowerSpectralDensitySummary,
PowerSpectralDensityEstimator,
ContingentNegativeVariation,
TimeLockedTopography,
TimeLockedContrast)
fname = './data/JSXXX-epo.fif'
if not op.exists(fname):
print('File not present, downloading...')
import urllib.request
url = 'https://ndownloader.figshare.com/files/13179518'
urllib.request.urlretrieve(url, fname)
print('Download complete')
epochs = mne.read_epochs(fname, preload=True)
psds_params = dict(n_fft=4096, n_overlap=100, n_jobs='auto', nperseg=128)
base_psd = PowerSpectralDensityEstimator(
psd_method='welch', tmin=None, tmax=0.6, fmin=1., fmax=45.,
psd_params=psds_params, comment='default')
# Note that the psd is shared by all `PowerSpectralDensity` markers.
# To save time, the PSD will not be re-computed.
# When making another set of marker, also recompute the base_psd explicitly.
m_list = [
PowerSpectralDensity(estimator=base_psd, fmin=1., fmax=4.,
normalize=False, comment='delta'),
PowerSpectralDensity(estimator=base_psd, fmin=1., fmax=4.,
normalize=True, comment='deltan'),
PowerSpectralDensity(estimator=base_psd, fmin=4., fmax=8.,
normalize=False, comment='theta'),
PowerSpectralDensity(estimator=base_psd, fmin=4., fmax=8.,
normalize=True, comment='thetan'),
PowerSpectralDensity(estimator=base_psd, fmin=8., fmax=12.,
normalize=False, comment='alpha'),
PowerSpectralDensity(estimator=base_psd, fmin=8., fmax=12.,
normalize=True, comment='alphan'),
PowerSpectralDensity(estimator=base_psd, fmin=12., fmax=30.,
normalize=False, comment='beta'),
PowerSpectralDensity(estimator=base_psd, fmin=12., fmax=30.,
normalize=True, comment='betan'),
PowerSpectralDensity(estimator=base_psd, fmin=30., fmax=45.,
normalize=False, comment='gamma'),
PowerSpectralDensity(estimator=base_psd, fmin=30., fmax=45.,
normalize=True, comment='gamman'),
PowerSpectralDensity(estimator=base_psd, fmin=1., fmax=45.,
normalize=True, comment='summary_se'),
PowerSpectralDensitySummary(estimator=base_psd, fmin=1., fmax=45.,
percentile=.5, comment='summary_msf'),
PowerSpectralDensitySummary(estimator=base_psd, fmin=1., fmax=45.,
percentile=.9, comment='summary_sef90'),
PowerSpectralDensitySummary(estimator=base_psd, fmin=1., fmax=45.,
percentile=.95, comment='summary_sef95'),
PermutationEntropy(tmin=None, tmax=0.6, backend='c'),
SymbolicMutualInformation(
tmin=None, tmax=0.6, method='weighted', backend='openmp',
method_params={'nthreads': 'auto'}, comment='weighted'),
KolmogorovComplexity(tmin=None, tmax=0.6, backend='openmp',
method_params={'nthreads': 'auto'}),
# Evokeds
ContingentNegativeVariation(tmin=-0.004, tmax=0.596),
TimeLockedTopography(tmin=0.064, tmax=0.112, comment='p1'),
TimeLockedTopography(tmin=0.876, tmax=0.936, comment='p3a'),
TimeLockedTopography(tmin=0.996, tmax=1.196, comment='p3b'),
TimeLockedContrast(tmin=None, tmax=None, condition_a='LSGS',
condition_b='LDGD', comment='LSGS-LDGD'),
TimeLockedContrast(tmin=None, tmax=None, condition_a='LSGD',
condition_b='LDGS', comment='LSGD-LDGS'),
TimeLockedContrast(tmin=None, tmax=None, condition_a=['LDGS', 'LDGD'],
condition_b=['LSGS', 'LSGD'], comment='LD-LS'),
TimeLockedContrast(tmin=0.736, tmax=0.788, condition_a=['LDGS', 'LDGD'],
condition_b=['LSGS', 'LSGD'], comment='mmn'),
TimeLockedContrast(tmin=0.876, tmax=0.936, condition_a=['LDGS', 'LDGD'],
condition_b=['LSGS', 'LSGD'], comment='p3a'),
TimeLockedContrast(tmin=None, tmax=None, condition_a=['LSGD', 'LDGD'],
condition_b=['LSGS', 'LDGS'], comment='GD-GS'),
TimeLockedContrast(tmin=0.996, tmax=1.196, condition_a=['LSGD', 'LDGD'],
condition_b=['LSGS', 'LDGS'], comment='p3b')
]
mc = Markers(m_list)
mc.fit(epochs)
mc.save('data/JSXXX-markers.hdf5', overwrite=True)
Out:
Reading ./data/JSXXX-epo.fif ...
Read a total of 2 projection items:
Average EEG reference (1 x 250) active
Average EEG reference (1 x 256) active
Found the data of interest:
t = -200.00 ... 1340.00 ms
0 CTF compensation matrices available
616 matching events found
Applying baseline correction (mode: mean)
Created an SSP operator (subspace dimension = 2)
616 matching events found
Applying baseline correction (mode: mean)
Not setting metadata
Created an SSP operator (subspace dimension = 2)
2 projection items activated
Fitting nice/marker/PowerSpectralDensity/delta
Autodetected number of jobs 8
Effective window size : 16.384 (s)
Fitting nice/marker/PowerSpectralDensity/deltan
Fitting nice/marker/PowerSpectralDensity/theta
Fitting nice/marker/PowerSpectralDensity/thetan
Fitting nice/marker/PowerSpectralDensity/alpha
Fitting nice/marker/PowerSpectralDensity/alphan
Fitting nice/marker/PowerSpectralDensity/beta
Fitting nice/marker/PowerSpectralDensity/betan
Fitting nice/marker/PowerSpectralDensity/gamma
Fitting nice/marker/PowerSpectralDensity/gamman
Fitting nice/marker/PowerSpectralDensity/summary_se
Fitting nice/marker/PowerSpectralDensitySummary/summary_msf
Fitting nice/marker/PowerSpectralDensitySummary/summary_sef90
Fitting nice/marker/PowerSpectralDensitySummary/summary_sef95
Fitting nice/marker/PermutationEntropy/default
Filtering at 10.42 Hz
Fitting nice/marker/SymbolicMutualInformation/weighted
Autodetected number of jobs 2
Computing CSD
Using EGI 256 locations for CSD
Using 2 jobs
Filtering at 10.42 Hz
Autodetected number of threads 4
Fitting nice/marker/KolmogorovComplexity/default
Running KolmogorovComplexity
Autodetected number of threads 4
Elapsed time 2.3484690189361572 sec
Fitting nice/marker/ContingentNegativeVariation/default
Fitting nice/marker/TimeLockedTopography/p1
Fitting nice/marker/TimeLockedTopography/p3a
Fitting nice/marker/TimeLockedTopography/p3b
Fitting nice/marker/TimeLockedContrast/LSGS-LDGD
Fitting nice/marker/TimeLockedContrast/LSGD-LDGS
Fitting nice/marker/TimeLockedContrast/LD-LS
Fitting nice/marker/TimeLockedContrast/mmn
Fitting nice/marker/TimeLockedContrast/p3a
Fitting nice/marker/TimeLockedContrast/GD-GS
Fitting nice/marker/TimeLockedContrast/p3b
Writing channel info to HDF5 file
Writing PSDS Estimator to HDF5 file
Channel info already present in HDF5 file, will not be overwritten
Channel info already present in HDF5 file, will not be overwritten
PSDS Estimator already present in HDF5 file, will not be overwritten
Channel info already present in HDF5 file, will not be overwritten
PSDS Estimator already present in HDF5 file, will not be overwritten
Channel info already present in HDF5 file, will not be overwritten
PSDS Estimator already present in HDF5 file, will not be overwritten
Channel info already present in HDF5 file, will not be overwritten
PSDS Estimator already present in HDF5 file, will not be overwritten
Channel info already present in HDF5 file, will not be overwritten
PSDS Estimator already present in HDF5 file, will not be overwritten
Channel info already present in HDF5 file, will not be overwritten
PSDS Estimator already present in HDF5 file, will not be overwritten
Channel info already present in HDF5 file, will not be overwritten
PSDS Estimator already present in HDF5 file, will not be overwritten
Channel info already present in HDF5 file, will not be overwritten
PSDS Estimator already present in HDF5 file, will not be overwritten
Channel info already present in HDF5 file, will not be overwritten
PSDS Estimator already present in HDF5 file, will not be overwritten
Channel info already present in HDF5 file, will not be overwritten
PSDS Estimator already present in HDF5 file, will not be overwritten
Channel info already present in HDF5 file, will not be overwritten
PSDS Estimator already present in HDF5 file, will not be overwritten
Channel info already present in HDF5 file, will not be overwritten
PSDS Estimator already present in HDF5 file, will not be overwritten
Channel info already present in HDF5 file, will not be overwritten
PSDS Estimator already present in HDF5 file, will not be overwritten
Channel info already present in HDF5 file, will not be overwritten
Channel info already present in HDF5 file, will not be overwritten
Channel info already present in HDF5 file, will not be overwritten
Channel info already present in HDF5 file, will not be overwritten
Channel info already present in HDF5 file, will not be overwritten
Writing epochs to HDF5 file
Channel info already present in HDF5 file, will not be overwritten
Epochs already present in HDF5 file, will not be overwritten
Channel info already present in HDF5 file, will not be overwritten
Epochs already present in HDF5 file, will not be overwritten
Channel info already present in HDF5 file, will not be overwritten
Epochs already present in HDF5 file, will not be overwritten
Channel info already present in HDF5 file, will not be overwritten
Epochs already present in HDF5 file, will not be overwritten
Channel info already present in HDF5 file, will not be overwritten
Epochs already present in HDF5 file, will not be overwritten
Channel info already present in HDF5 file, will not be overwritten
Epochs already present in HDF5 file, will not be overwritten
Channel info already present in HDF5 file, will not be overwritten
Epochs already present in HDF5 file, will not be overwritten
Channel info already present in HDF5 file, will not be overwritten
Epochs already present in HDF5 file, will not be overwritten
Channel info already present in HDF5 file, will not be overwritten
Epochs already present in HDF5 file, will not be overwritten
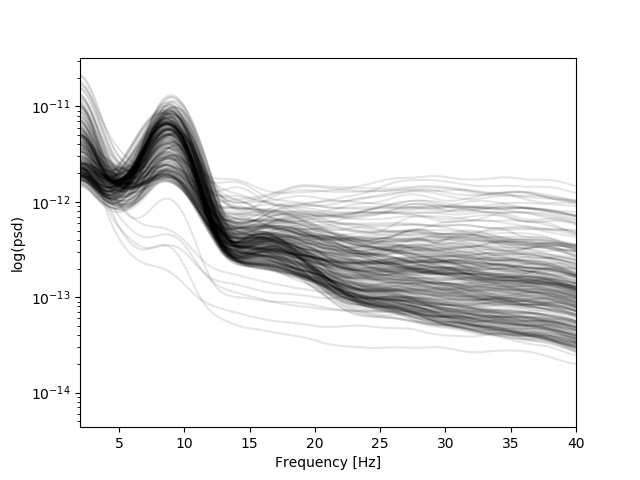
Let’s explore a bit the PSDs used for the marker computation
psd = base_psd.data_
freqs = base_psd.freqs_
plt.figure()
plt.semilogy(freqs, np.mean(psd, axis=0).T, alpha=0.1, color='black')
plt.xlim(2, 40)
plt.ylabel('log(psd)')
plt.xlabel('Frequency [Hz]')
plt.show()
# We clearly see alpha and beta band peaks.
Total running time of the script: ( 2 minutes 5.099 seconds)